Monte Carlo Cross Validation (MCCV) is a machine learning framework designed to estimate clinical outcomes and effects in patient cohorts to generate clinical and biological hypotheses. MCCV was used to in the following publications:
- Giangreco NP, Lebreton G, Restaino S, Jane Farr M, Zorn E, Colombo PC, Patel J, Levine R, Truby L, Soni RK, Leprince P, Kobashigawa J, Tatonetti NP, Fine BM. Plasma kallikrein predicts primary graft dysfunction after heart transplant. J Heart Lung Transplant. 2021 Jul 10:S1053-2498(21)02391-3. doi: 10.1016/j.healun.2021.07.001.
- Giangreco, N.P., Lebreton, G., Restaino, S. et al. Alterations in the kallikrein-kinin system predict death after heart transplant. Sci Rep 12, 14167 (2022). https://doi.org/10.1038/s41598-022-18573-2
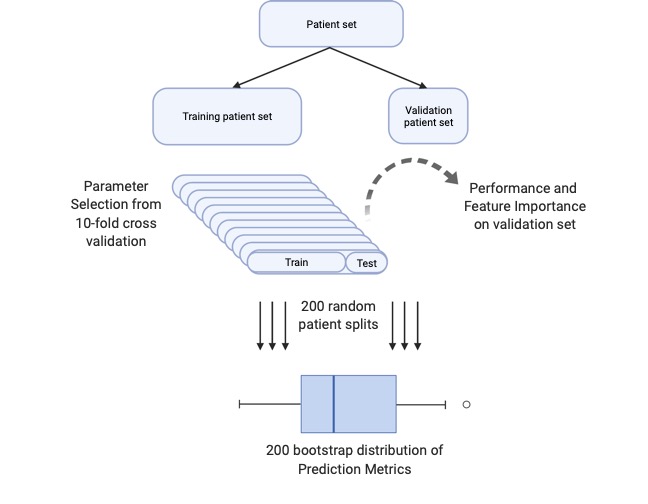
The prediction scheme, Monte Carlo Cross Validation (MCCV), is composed of 5 steps repeated multiple times (e.g. N=200):
- Split the data into 85% training and 15% validation sets.
- Separately normalize, or subtract the sample mean and divide by the sample standard deviation, the training and testing data.
- Using only the sampled training data, compute ten-fold cross validation and choose the top performing model parameters for predicting the response.
- Refit the training dataset using the top-prediction model parameters determined in step 3.
- Predict patient response in the yet-to-be-seen validation set using the refit model calculated in step 4.
The above publications resulted from my PhD work at Columbia University. In my free time after my PhD, I put together the python functions and scripts as a package for MCCV routines. Additionally, I am conducting a simulation experiment to evaluate outcome prediction for different biomarker distributions.
MCCV (Shiny for Python) App
A web application to simulate prior data distributions and inspective predictive evidence using MCCV
python
shiny
app
shiny-for-python
prediction
MCCV for Biomarker Prediction
python
biomarker
distributions
simulation
prediction
differential expression
No matching items